FandPLimitTool
Software [1] for computing single molecule localization accuracies and resolution measures
Parameter estimation problems play a central role in single molecule image analysis. Examples include determination of the 2D location of single molecules [2], estimating the level of defocus of single molecules [3] [4] [5], determining the distance of separation between two single molecules [6] [7] [8], estimation of photon count [9], etc. Common to all these problems is that it is helpful for the experimenter to have tools available that can quantify the accuracy with which various parameters can be estimated.
The FandPLimitTool [1] is a GUI based software module that allows users to calculate the limits to the accuracy with which parameters can be estimated from single molecule imaging data. The software supports calculation of limits for the 2D/3D location estimation problem and the 2D/3D distance-estimation/resolution problem. The location estimation problem is concerned with the task of determining the position of a single molecule and the distance-estimation/resolution problem is concerned with the task of determining the distance of separation between two single molecules. The user can calculate limits for a variety of imaging scenarios.
YouTube Tutorial (Part I: Localization Accuracy):
YouTube Tutorial (Part II: Resolution Limit):
FandPLimitTool main window together with required parameters window, multivalue mode window and plots.
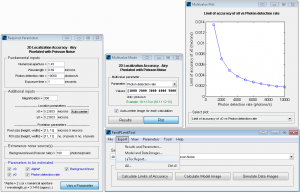
FandPLimitTool advanced parameters window.
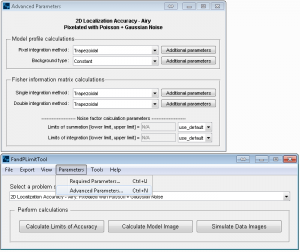
FandPLimitTool summary and console windows.
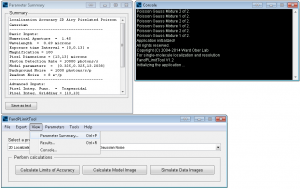
FandPLimitTool model and data image viewer windows.
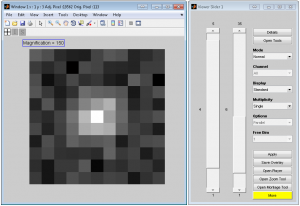
The technical details of the mathematical framework used for calculating the accuracy limits can be found in [9]. Accuracy limits for the location estimation problem can be found in [3] [4] [5]. Accuracy limit for the separation distance estimation problem can be found in [6] [7] [8].
The software is written in MATLAB and designed using the object oriented analysis and design methodology. The software has been designed to be extensible to allow for new models and numerical calculation routines.
References
[1] A. V. Abraham, S. Ram, J. Chao, E. S. Ward, and R. J. Ober, Quantitative study of single molecule location estimation techniques. Opt. Express, 17:23352-23373, 2009. cite for the software (FandPLimitTool)
[2] R. J. Ober, S. Ram, and E. S.Ward. Localization accuracy in single molecule microscopy.Biophys. J., 86:1185-1200, 2004. cite for 2D localization accuracy
[3] S. Ram, R. J. Ober, and E. S. Ward. How accurately can a single molecule be localized in three dimensions when imaged through a fluorescence microscope? Proc. of SPIE, 5699:426-435, 2005. cite for 3D localization accuracy for a conventional microscopy setup
[4] S. Ram, J. Chao, P. Prabhat, E. S. Ward, and R. J. Ober. A novel approach to determining the three-dimensional location of microscopic objects with applications to 3D particle tracking. Proc. of SPIE, 6443:64430C, 2007. cite for 3D localization accuracy for a conventional and/or a multifocal plane microscopy (MUM) setup
[5] S. Ram, P. Prabhat, J. Chao, E. S. Ward, and R. J. Ober. High accuracy 3D quantum dot tracking with multifocal plane microscopy for the study of fast intracellular dynamics in live cells. Biophys. J., 95:6025-6043, 2008. cite for 3D localization accuracy for a conventional and/or multifocal plane microscopy (MUM) setup
[6] S. Ram, E. S. Ward, and R. J. Ober. Beyond Rayleigh’s criterion: A resolution measure with application to single-molecule microscopy. Proc. Natl. Acad. Sci. USA, 103:4457-4462, 2006. cite for 2D resolution
[7] S. Ram, E. S. Ward, and R. J. Ober. A novel resolution measure for optical microscopes: Stochastic analysis of the performance limits. Proceedings of the 2006 IEEE International Symposium on Biomedical Imaging: Macro to Nano, 770-773, 2006. cite for 2D resolution
[8] J. Chao, S. Ram, A. V. Abraham, E. S. Ward, and R. J. Ober, A resolution measure for three-dimensional microscopy. Opt. Commun., 282:1751-1761, 2009. cite for 3D resolution
[9] S. Ram, E. S. Ward, and R. J. Ober. A stochastic analysis of performance limits for optical microscopes. Multidim. Syst. Sign. Process., 17:27-57, 2006. cite for the mathematical framework for the calculation of limits of accuracy
[10] J. Chao, E. S.Ward, and R. J. Ober, Fisher information matrix for branching processes with application to electron-multiplying charge-coupled devices. Multidim. Syst. Sign. Process., 23:349-379, 2012. cite for EMCCD-based calculations
[11] J. Chao, S. Ram, E. S. Ward, and R. J. Ober, Ultrahigh accuracy imaging modality for super-localization microscopy. Nat. Methods, 10:335-338, 2013. cite for EMCCD-based calculations
[12] A. Tahmasbi, S. Ram, J. Chao, A. V. Abraham, F. Tang, E. S. Ward, and R. J. Ober, Designing the focal plane spacing for multifocal plane microscopy. Opt. Express, 22:16706-16721, 2014. cite for the MUMDesignTool
Version History
- V1.2 (June 30, 2014) This is the currently available version.
- V1.1 (May 8, 2008) Please note that this is a Beta version.
- V1.0 (2006)
Minimum System Requirements
- PC with Core 2 Duo or equivalent processor suggested
- Windows XP Service Pack 2 *
- 512 MB RAM
- 1024 x 768 screen resolution
* Although MATLAB and the MCR are available on the Linux/Unix and MacOS platforms, this executable version of the software can run only on a PC with Windows XP, Windows Server 2008, Windows 7 and Windows 8, and will NOT work in a Linux/Unix or Mac environment.
How to obtain the software
If you are interested in access to one of our software, please contact us at info@wardoberlab.com
Download FandPLimitTool User’s Manual 
Download FandPLimitTool License Agreement 
FAQ
Q: Why is starting the software for the first time after the Windows OS startup slow?
A: The initialization of the FandPLimitTool after the Windows OS startup may take up to a minute, depending on the computer’s specifications, due to the time required for loading the MATLAB MCR files from the hard drive into the memory. This time will be significantly reduced (to often several seconds) in the subsequent runs. During this initialization step, the following splash window informs the user about the progress.
FandPLimitTool splash window.

Release Notes
The newly released FandPLimitTool V1.2 is loaded with new features including:
- A new software module the MUMDesignTool is incorporated with the package which provides tools to design the focal plane spacing for multifocal plane microscopy.
- New functionality that allows the calculation of the 3D resolution measure.
- A new function called “vary a parameter” is added to the software which allows users to calculate and plot the limits of accuracy for a range of different experimental parameters.
- New functionality that allows simulating single molecule model and data images.
- New functionality that allows determining limits of accuracy in the presence of stochastic signal amplification, i.e., when using an electron-multiplying CCD (EMCCD) detector.
- The code is significantly optimized. The software therefore runs much faster than the previous version.
- Exporting tools are enhanced and now user can export the results, parameters, simulated model and data images and graphs as text, image and LaTex report files, respectively.
- A console window and a log file to facilitate bug reporting and enhanced user interface design.
Bug Reporting
We are constantly trying to improve this software and would be very grateful if you could send us a report on any bugs you find. Please send all bug reports to the email address software@wardoberlab.com with the subject FandPLimitTool Bug. Please include as many details about the bug as you can, including details of all tasks that led up to exposure of the bug. This will help us in finding a solution to the problem. When we have a fix for the problem, we will release an updated version of the software. Please look in the Release Notes section for problems we are already aware of and have solutions for.
FandPLimitTool Development Team
Current Members
- Raimund J. Ober (Team Leader)
- Anish V. Abraham
- Jerry Chao
- E. Sally Ward
Past Members
- Amir Tahmasbi
- Sripad Ram
Acknowledgments
The development work for FandPLimitTool was supported in part by the National Institutes of Health.
Contact Address
Raimund J. Ober, Ph.D.Professor |
E. Sally Ward, Ph.D.Professor |
